


Next: 2.6 Remove <parameter>
Up: 2 Commands
Previous: 2.4 Fit
Once you have fitted the model you should inspect the
residual variation around the predicted values. The program
calculates the following variables:
- pi
- The frequencies of the donor allele predicted
by the fitted model.
- zi
- Standarized residuals,

The deviance between observed frequencies and the predicted
values divided by the binomial variance around the predicted
value under the fitted model.
- gi
- The sum of the contribution of all plant
varieties k=1 and k=2 to plant i,
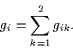
See [1] equation (6). This variable may be a
useful predictor of self-fertilization.
These new variables are written to <filename> to the
right of the original data variables (see section
2.1). You should import this file into you
favorite plotting package and plot the residuals against
several of the other variables, e.g. the distance and
direction from the donor-field.
![]()
![]()
![]()
![]()