


Next: 2 The model
Up: Inferring patterns of migration
Previous: Contents
Contents
In population genetics, there has long been an interest in
understanding and analyzing the gene frequency structure
found in natural populations. Local genetic drift will tend
to create genetic differences between different subunits of
a population. These differences will, after a number of
initial generations, be balanced by migration. One can thus,
in principle, estimate the amount of migration between the
different subunits from the observed gene frequencies. Most
earlier approaches to this problem have, in part, relied on
the assumptions of the island model. Under that model
one can estimate the number of migrants 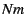 from the
estimated amount of genetic differentiation as measured by
e.g. Wright's parameter
from the
estimated amount of genetic differentiation as measured by
e.g. Wright's parameter 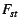 . Typically, these
estimates are then, in further exploratory analysis,
regressed against geographic distances to test for isolation
by distance effects predicted by some idealized theoretical
models. This manual documents the use of some S-Plus and c
code I wrote to implement an alternative solution of this
problem based more directly on standard statistical
principles, which allows much less restrictive assumptions
about the underlying pattern of migration. The general
method, which is based on an idea in Felsenstein (1982),
is described in two papers (Tufto et al., 1996; Tufto and Hindar, 2002; Tufto et al., 1998); preprints
of these are available on my list of
publications. A brief
summary of the approach is also given here.
. Typically, these
estimates are then, in further exploratory analysis,
regressed against geographic distances to test for isolation
by distance effects predicted by some idealized theoretical
models. This manual documents the use of some S-Plus and c
code I wrote to implement an alternative solution of this
problem based more directly on standard statistical
principles, which allows much less restrictive assumptions
about the underlying pattern of migration. The general
method, which is based on an idea in Felsenstein (1982),
is described in two papers (Tufto et al., 1996; Tufto and Hindar, 2002; Tufto et al., 1998); preprints
of these are available on my list of
publications. A brief
summary of the approach is also given here.
Readers who prefer to read a printed version of this
manual can download the .pdf version of the manual
from this
url.
It is assumed that the reader has access to and some
familiarity with R or S-plus.



Next: 2 The model
Up: Inferring patterns of migration
Previous: Contents
Contents
Jarle Tufto
2001-08-28